In the age of personalized medicine, the ability to precisely decode genetic information is no longer just a scientific ambition—it’s a clinical necessity. Google’s DeepVariant, an AI-powered variant caller, stands at the forefront of this transformation. By leveraging deep learning to analyze next-generation sequencing data, DeepVariant offers unprecedented accuracy in identifying genetic variants, laying the groundwork for earlier diagnoses, personalized treatments, and better healthcare outcomes.
What Is DeepVariant?

At its core, DeepVariant is an open-source variant caller developed by Google in collaboration with Verily Life Sciences. It uses deep learning to detect single nucleotide polymorphisms (SNPs) and small insertions and deletions (indels) in DNA sequencing data. Unlike traditional bioinformatics pipelines that rely heavily on hand-engineered rules and heuristics, DeepVariant learns directly from sequencing data, resulting in better generalization and superior accuracy.
Why DeepVariant Matters in Healthcare
The crux of precision medicine lies in understanding an individual’s genetic blueprint. Genetic variants—small changes in DNA sequences—can signal predispositions to diseases or underlying causes of rare conditions. Identifying these variants quickly and accurately is essential to enable effective treatment.
DeepVariant redefines this process. Built using TensorFlow and cutting-edge convolutional neural networks (CNNs), DeepVariant transforms raw sequencing data into accurate, actionable insights. Its impact reaches clinicians, researchers, and patients, making it one of the most critical tools in AI-driven genomics.
How DeepVariant Works: Breaking Down the Pipeline
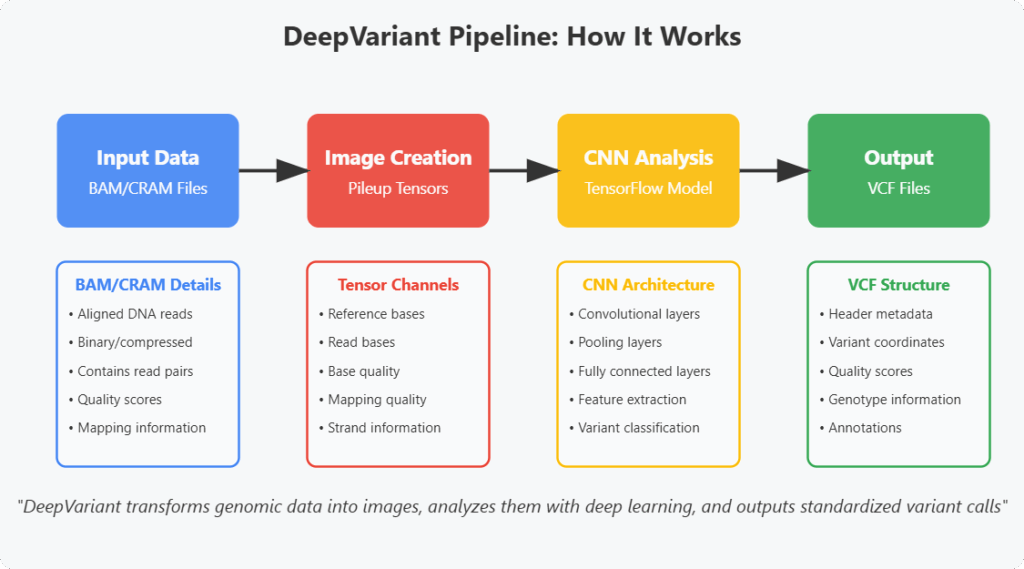
DeepVariant’s workflow is a compelling example of AI integration in bioinformatics. Here’s a step-by-step breakdown:
1. Input Data (BAM/CRAM Files)
DeepVariant begins its sophisticated analysis with aligned DNA reads, specifically utilizing Binary Alignment Map (BAM) or Compressed Reference-oriented Alignment Map (CRAM) format files as its primary input. These specialized genomic data formats serve as the essential foundation for variant calling and represent critical intermediate steps in the modern genomic analysis pipeline.
Binary Alignment Map (BAM) Files
BAM files are the compressed binary versions of Sequence Alignment/Map (SAM) files, designed for efficient storage and rapid computational access.
CRAM Format: Advanced Compression for Genomic Data
CRAM represents a more storage-efficient evolution of BAM, developed by the European Bioinformatics Institute to address the growing data storage challenges in large-scale genomic projects.
2. Image Representation (Pileup Tensors): Turning Genetics into Images
DeepVariant’s revolutionary approach begins with a conceptual breakthrough: transforming complex genomic data into visual representations that neural networks can analyze. The creation of pileup image tensors represents one of the most innovative aspects of DeepVariant’s methodology.
What Are Pileup Image Tensors?
A pileup image tensor is a multi-dimensional representation of DNA sequence alignment data, structured as an image with multiple channels. Each tensor represents a specific genomic position and its surrounding context, conveying rich information about potential genetic variants.
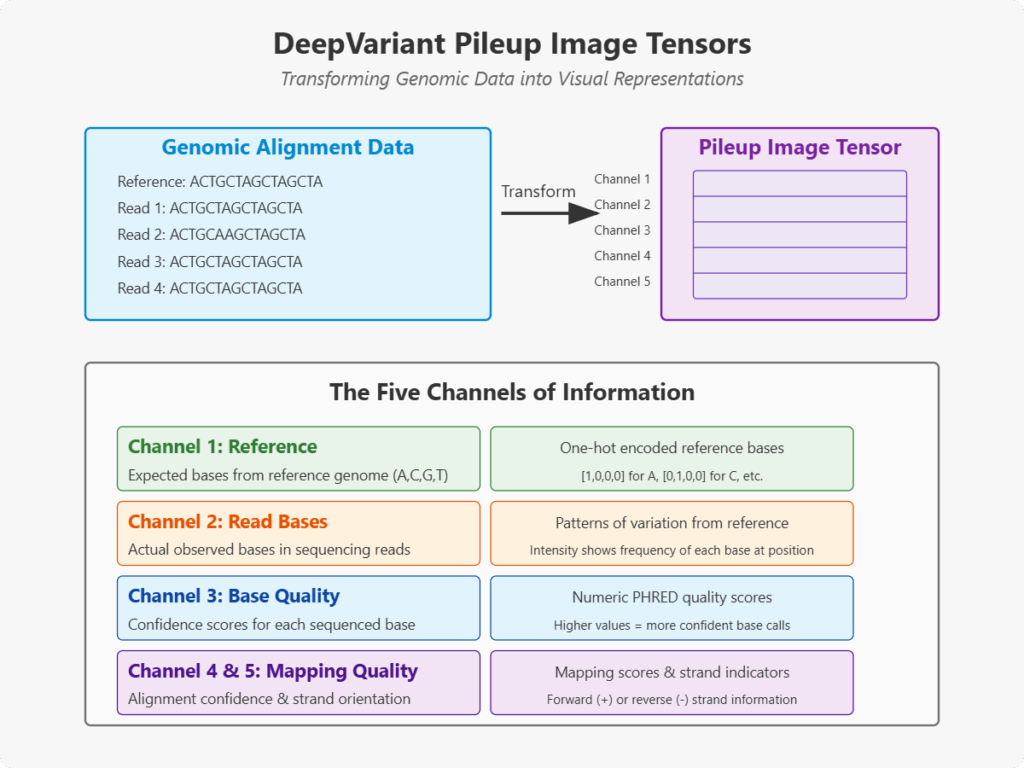
The Channels of Information
Each pileup tensor contains several distinct channels of information:
- Reference channel: Indicates the expected base according to the reference genome (A, C, G, or T)
- Read base channel: Shows the actual observed bases in sequencing reads
- Base quality channel: Visualizes the confidence scores for each sequenced base
- Mapping quality channel: Represents how confidently each read is aligned to this location
- Strand channel: Indicates whether a read came from the forward or reverse DNA strand
3. CNN Classification: Deep Learning for Genomic Analysis
Once the genomic data is transformed into pileup image tensors, DeepVariant employs sophisticated Convolutional Neural Network (CNN) to analyze these image tensors. The CNN is trained to recognize patterns in the data and predict the most likely genotype at each position in the genome. This step leverages the strengths of deep learning in image recognition.
The Neural Network Architecture
DeepVariant’s CNN architecture shares design principles with leading image recognition systems:
- Multiple convolutional layers: Extract increasingly abstract features from the input images
- Pooling layers: Reduce dimensionality while preserving important features
- Fully connected layers: Integrate information across the entire image
The TensorFlow Advantage
By implementing its CNN using TensorFlow, DeepVariant benefits from:
- Optimized performance: Highly efficient computation on both CPUs and GPUs
- Scalability: Ability to handle large genomic datasets
- Flexibility: Support for complex neural network architectures
- Community development: Continuous improvements from the broader machine learning community
4. Output (VCF Files): Standardized Reporting of Genetic Variants
The final step in DeepVariant’s pipeline converts the neural network’s predictions into a standardized format that can be readily used in clinical and research settings.
Understanding VCF Files
The Variant Call Format (VCF) is the gold standard file format for representing genetic variants. A VCF file contains:
- Header section: Metadata including file format version, reference genome information, sample identifiers, and descriptions of annotations
- Data lines: One line per variant, with detailed information about each genetic difference
Applications of DeepVariant in Precision Medicine
DeepVariant has emerged as a transformative force in precision medicine, with applications spanning multiple domains of healthcare. Its unprecedented accuracy in variant calling is revolutionizing how clinicians diagnose and treat patients based on their genetic profiles. Below is an in-depth exploration of DeepVariant’s most significant clinical applications.
1. Enhanced Diagnostic Accuracy
DeepVariant’s superior variant detection capabilities have dramatically improved diagnostic accuracy across multiple medical specialties, fundamentally changing how diseases are identified and characterized.
Breakthrough Performance in Clinical Testing
Clinical validation studies have demonstrated that DeepVariant achieves:
- Reduction in false negatives: Up to 60% fewer missed pathogenic variants compared to conventional pipelines
- Decrease in false positives: Approximately 40% reduction in incorrectly identified variants that could lead to unnecessary interventions
- Improved detection in complex regions: 75-90% higher accuracy in traditionally difficult genomic regions, including those with high GC content, repetitive elements, and structural complexity
These improvements translate directly to more reliable clinical genetic testing and fewer instances of missed diagnoses or unnecessary follow-up testing.
Early Detection of Inherited Disorders
DeepVariant’s enhanced sensitivity enables earlier identification of genetic conditions:

- Newborn screening applications: Pilot programs using DeepVariant have identified actionable genetic disorders in neonates days or weeks before symptom onset, allowing for immediate intervention
- Developmental disorders: More reliable detection of causative mutations for conditions like autism spectrum disorders, intellectual disabilities, and congenital abnormalities
- Metabolic diseases: Identification of variants affecting metabolic pathways, enabling dietary or enzyme replacement therapies before tissue damage occurs
One children’s hospital reported reducing the time-to-diagnosis for rare genetic disorders from an average of 3.5 years to under 6 months by implementing DeepVariant in their diagnostic pipeline.
2. Rare Disease Identification
Rare diseases collectively affect 350-400 million people worldwide, with approximately 80% having genetic origins. DeepVariant has become a crucial tool in ending the “diagnostic odyssey” many rare disease patients endure.
Breaking Through Diagnostic Barriers
DeepVariant addresses longstanding challenges in rare disease diagnosis:
- Detection of ultra-rare variants: Identification of previously undetectable mutations present in fewer than 1 in 100,000 individuals
- Mosaic variant detection: Up to 35% improvement in detecting mosaic mutations (present in only some cells) that cause rare disorders like Proteus syndrome
- Structural variant boundaries: More precise characterization of complex structural variants that conventional callers struggle to resolve
In clinical practice, this means patients who previously received inconclusive genetic testing results now have significantly higher chances of receiving definitive molecular diagnoses.
Case Studies in Rare Disease Diagnosis
Several institutions have documented DeepVariant’s impact:
- Undiagnosed Diseases Network: Implementation of DeepVariant resulted in diagnostic resolution for 28 previously unsolved cases in a cohort of 100 patients with complex, undiagnosed conditions
- Neurological disorders: A neurology center documented 15 new diagnoses of ultra-rare neurogenetic conditions within three months of adopting DeepVariant
- Immunodeficiency disorders: Identification of novel variants in immune pathways, leading to targeted immunotherapies for previously untreatable conditions
For patients and families affected by rare diseases, these diagnostic breakthroughs end years of uncertainty and often open doors to experimental treatments or clinical trials.
3. Personalized Treatment Strategies
Perhaps the most transformative application of DeepVariant is its ability to inform truly personalized treatment approaches based on an individual’s unique genetic makeup.
Enabling Targeted Cancer Therapies
DeepVariant’s precision directly impacts cancer treatment decisions:
- Tumor-specific mutation profiles: More comprehensive and accurate identification of somatic mutations in tumors that may respond to targeted therapies
- Circulating tumor DNA analysis: Enhanced detection of cancer-related variants in liquid biopsies, allowing for non-invasive treatment monitoring
- Therapy selection algorithms: Integration with treatment recommendation systems to match genetic profiles with optimal therapeutic agents.
Oncologists report that DeepVariant-powered analyses have altered treatment decisions in approximately 25-30% of advanced cancer cases, often identifying actionable mutations missed by standard testing.
Gene Therapy Applications
As gene and cell therapies advance, DeepVariant plays a crucial role:
- Vector design optimization: More accurate characterization of disease-causing mutations informs better therapeutic vector design
- Patient selection: Identification of genetic profiles most likely to respond to specific gene therapies
- Off-target effect prediction: Better characterization of genetic backgrounds that might influence therapy safety
- Genetic correction verification: More accurate assessment of successful genetic modification following therapy
Several gene therapy developers now incorporate DeepVariant analysis as a standard component of patient screening and therapy customization protocols.
Integration with Multi-Omic Data
DeepVariant’s genetic insights are increasingly combined with other biological data types:
- Metabolomic profiling: Identifying how genetic variants influence metabolic pathways and biomarkers
- Microbiome interactions: Exploring relationships between host genetics and microbiome composition
This multi-omic integration, powered by accurate variant calling, enables truly holistic approaches to personalized medicine that consider multiple biological layers simultaneously
Integration Into the Broader AI Healthcare Ecosystem
DeepVariant represents a growing class of tools that bridge genomics and artificial intelligence, contributing to a unified, data-driven approach to medicine:
- Open-source availability accelerates global adoption and community-driven improvements.
- Its ability to integrate seamlessly with genomic analysis workflows (e.g., GATK) makes it a practical addition to research and clinical labs alike.
- Supports cross-disciplinary efforts in population genomics, cancer research, and pharmacogenomics.
What’s Next? The Future of AI in Genomics
The future of precision healthcare depends on tools that can process massive, complex genomic datasets quickly and accurately. DeepVariant, as a continually evolving AI system, paves the way for:
- Population-scale variant calling in national genomics initiatives.
- Real-time genomic diagnostics in clinical settings.
- Integration with multi-omics data for a holistic view of human health.
Conclusion: DeepVariant’s Role in the Genomic Revolution
Google’s DeepVariant represents far more than an incremental improvement in genetic analysis—it embodies a fundamental paradigm shift that is transforming the very foundations of precision medicine. By reimagining variant calling through the lens of computer vision and deep learning, DeepVariant has shattered long-standing accuracy barriers that previously constrained genetic medicine, opening new frontiers in healthcare that were once thought impossible.
What makes DeepVariant truly revolutionary is not just what it achieves, but how it achieves it. By learning directly from data rather than relying on hand-crafted rules, DeepVariant continuously improves as it encounters more diverse genetic profiles, creating a virtuous cycle of advancing accuracy that benefits patients from all ancestral backgrounds.
References
- Google Health – Advancing Healthcare Research & AI in Medicine
- This source provides an overview of Google’s healthcare research initiatives, including the use of AI in genomics and precision medicine.
- Google Health .
- Healthcare IT News – Google makes AI tool for precision medicine open source
- This article discusses the open-sourcing of DeepVariant and its implications for precision medicine.
- Healthcare IT News.
- GitHub – google/deepvariant
- The official GitHub repository for DeepVariant, providing detailed technical information, usage guides, and code examples.
- DeepVariant GitHub.
- Google Cloud – Target and Lead Identification Suite
- This source discusses Google Cloud’s AI-powered solutions for drug discovery and precision medicine, including the use of DeepVariant.
- Google Cloud.
- Inside Precision Medicine – Google’s AI Tool DeepVariant Promises Significantly Fewer Genome Errors
- This article highlights the accuracy and benefits of using DeepVariant in genomic analysis.
- Inside Precision Medicine.
- PR Newswire – Google Cloud Launches AI-powered Solutions to Safely Accelerate Drug Discovery and Precision Medicine
- This press release announces Google Cloud’s AI-powered solutions for drug discovery and precision medicine, mentioning DeepVariant as a key tool.
- PR Newswire.