
In this blog, we explore how artificial intelligence can integrate different types of biomedical data to revolutionize drug discovery, told through the fictional journey of Dr. Ananya.
Setting the Stage: A Researcher’s Journey
Dr. Ananya, a computational biologist, was working late in her lab, reviewing the results of her latest experiment. Once again, a molecule that looked good on paper had failed in biological tests. She had used advanced chemical analysis tools, but they couldn’t explain why the compound didn’t work in real cells. Frustrated, she sighed, “I need more than just chemical structure.” That moment sparked her interest in a new approach: using AI to combine data from genes, proteins, chemicals, and patient records to gain a deeper understanding of how drugs work — integrating genomic, chemical, proteomic, and clinical data to rethink how drugs are discovered.
Table of Contents
- Why Drug Discovery Needs AI and Multimodal Integration
- Understanding Small Molecule Representations
- SMILES (Simplified Molecular Input Line Entry System)
- Molecular Fingerprints
- Molecular Graphs
- 3D Coordinates
- How AI Powers Multimodal Drug Discovery
- AI Architectures: From Predictive Models to Generative Systems
- Learning Paradigms Empowering AI in Chemistry
- Self-Supervised Learning
- Reinforcement Learning
- Meta & Few-shot Learning
- Applications and Success Stories
- Virtual Screening
- Antibiotic Discovery
- Multi-objective Optimization
- Remaining Challenges
- What Lies Ahead: The Future of Multimodal AI in Drug Discovery
- Conclusion
- References
Why Drug Discovery Needs AI and Multimodal Integration
Fueled by curiosity, Dr. Ananya began to map out the complexity of the drug discovery landscape. She realized that predicting drug behavior requires more than chemical properties. It involves seeing how a drug interacts with the biological system at multiple levels.
Drug discovery is a lengthy, expensive, and highly complex process. On average, bringing a single drug to market takes more than 10 years and over $2.6 billion in investment. Moreover, the failure rate in clinical trials remains staggeringly high, with fewer than 10% of drug candidates ultimately approved for use (Deng et al., 2022).
This inefficiency largely stems from the intricate and nonlinear nature of biological systems. Drug responses depend on multiple layers of biological data: from genetic mutations and protein activity to metabolic pathways, chemical structure interactions, and patient-specific variables. Traditionally, these data types are analyzed separately, making it difficult to understand the full picture.
Multimodal data integration addresses this problem by combining diverse data sources into a unified framework, enabling a holistic view of biological systems. Artificial intelligence (AI), particularly Deep learning, has emerged as the most powerful tool for learning from and integrating these complex datasets.
Ananaya began exploring multimodal data integration — a way to combine diverse biological signals into a single framework. This approach could transform isolated data points into connected insights.
Understanding Small Molecule Representations
Before diving into how AI integrates multimodal data, Dr. Ananya realized she needed to understand how molecules are represented computationally. Just like sentences in language, molecules need a structured format for machines to process.
Here are the most common ways molecules are represented:
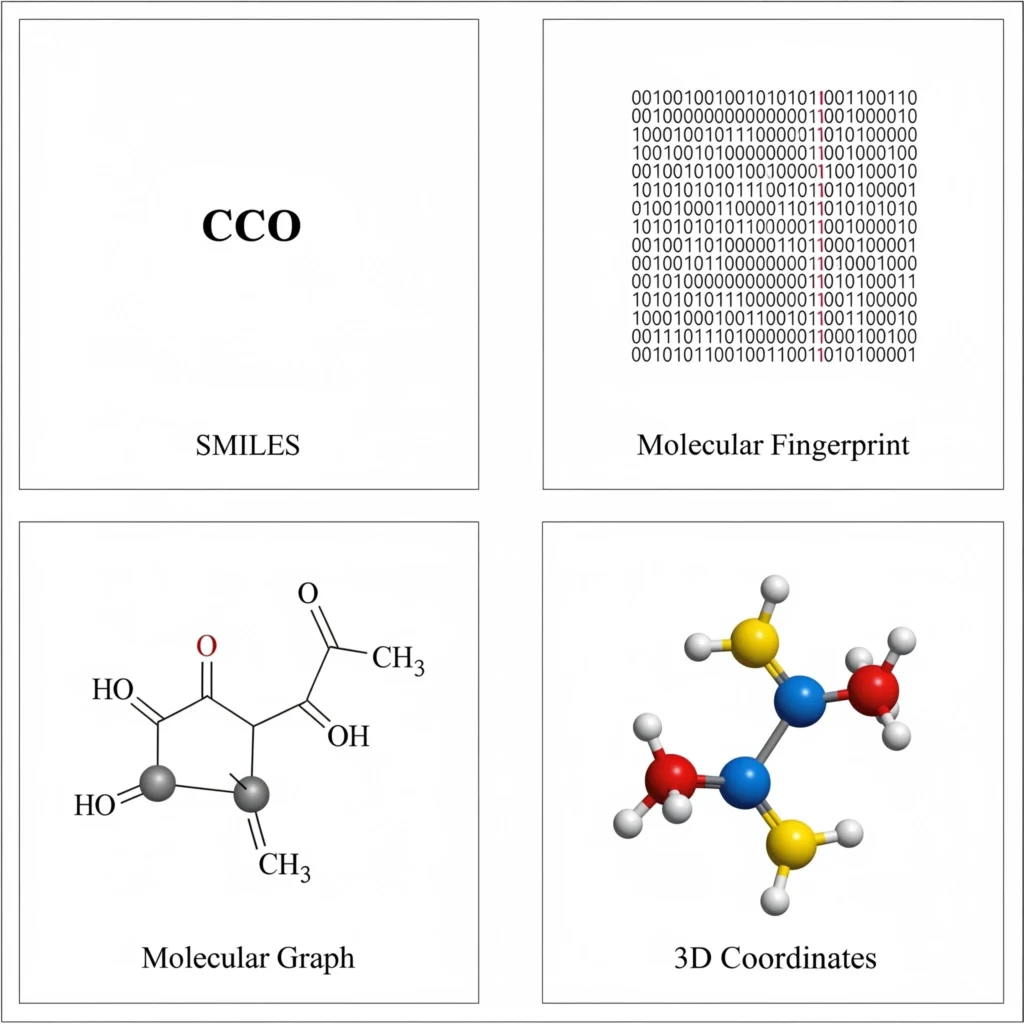
- SMILES (Simplified Molecular Input Line Entry System): A line of text that encodes a molecule’s structure. For example, ‘CCO’ represents ethanol. SMILES is widely used for its simplicity and compatibility with text-based models.
- Molecular Fingerprints: These are bit vectors that capture the presence of chemical substructures. They’re like molecular barcodes used for similarity searches and classification tasks.
- Molecular Graphs: A molecule can be viewed as a graph where atoms are nodes and bonds are edges. Graph Neural Networks (GNNs) use this structure to extract and learn relational information.
- 3D Coordinates: This format captures the physical spatial arrangement of atoms in 3D space and is vital for modeling binding affinity or docking.
Understanding these representations helped Dr. Ananya build a bridge from raw molecular data to the deep learning architectures she would later use. With this foundation, she was ready to explore how AI could learn meaningful insights across different data types — from molecular shape to biological response.
How AI Powers Multimodal Drug Discovery
Before she could design effective AI models, Dr. Ananya also had to understand the scope and quality of the data she was working with. She explored some of the most important chemical databases in the field:
- PubChem (by NIH): Contains over 111 million chemical structures and 271 million bioactivity data points from 750 sources (as of 2020). It’s a rich resource, but uncurated, which can introduce noise.
- ChEMBL (by EMBL): Offers curated data with over 1.6 million unique compounds and 14 million activity records. Frequently used in benchmarking.
- ZINC (by UCSF): A collection of over 120 million purchasable, annotated drug-like molecules. Subsets like ZINC-250k are widely used in AI training.
Each of these data sources contributes to building more robust, diverse, and informative AI models.
With her whiteboard filled with sketches of genomic sequences, protein pathways, and SMILES strings, Dr. Ananya began her experiments with AI models that could learn from all of them.
AI enhances drug discovery through two fundamental tasks:
- Molecular Property Prediction: Estimating properties such as toxicity, solubility, or binding affinity.
- Molecule Generation: Designing new drug-like compounds with desirable biological effects.
She began building a model that could learn from various types of biomedical data. Here’s what she had to integrate:
| Modality | Description | Example |
| Genomics | DNA sequence data | SNPs, mutations |
| Transcriptomics | RNA expression levels | mRNA levels from RNA-Seq |
| Proteomics | Protein abundance/function | Mass spectrometry data |
| Imaging | Visual scans of tissues/organs | MRI, pathology slides |
| Chemical Data | Molecular structure & bioactivity | SMILES, molecular graphs |
| Clinical Records | Patient history and diagnosis | EHRs, diagnoses, treatment records |
She soon realized that not all data is equal — integrating these modalities meant addressing missing values, aligning different formats, and creating consistent encodings. It was messy, but necessary.
AI Architectures: From Predictive Models to Generative Systems
Determined to go beyond shallow models, Dr. Ananya studied deep learning architectures used in modern drug discovery.
AI models in the drug discovery process molecules using various representations—fingerprints, SMILES strings, graphs, or 3D coordinates—and feed them into specialized neural networks. Each architecture is designed to suit the structure and complexity of a specific data type or task.
| Model Type | Use Case | Why It’s Used |
| CNNs | Image-based molecule analysis | Captures spatial features from 2D molecular images or protein-ligand maps |
| RNNs (LSTM/GRU) | SMILES-based molecule generation | Learns sequential dependencies in SMILES strings for decoding and synthesis |
| GNNs | Property prediction & generation | Understands the relational structure of atoms and bonds in a molecule |
| VAEs | Latent molecule design | Encodes molecules into a latent space for structured, interpretable generation |
| GANs | High-quality molecule synthesis | Trains a generator-discriminator pair to produce realistic and novel molecules |
| Transformers | Self-supervised learning on SMILES/graphs | Leverages attention mechanisms for better representation learning |
These models allow end-to-end learning, where features are learned directly from data—no manual descriptor engineering needed.
She experimented with Graph Neural Networks (GNNs), which represent molecules as graphs of atoms and bonds.
The model began predicting biological activity from structural data, but it still missed broader systemic effects. Her next move? Learning paradigms that would help the model think beyond one representation.

Learning Paradigms Empowering AI in Chemistry
Dr. Ananya explored advanced paradigms to build more generalizable models:
But Dr. Ananya knew that simply building models wasn’t enough. Her early GNN showed promise, but it lacked adaptability, especially when data was limited or noisy. She dove deeper into learning paradigms that could make her models more robust and transferable across tasks.
Self-Supervised Learning
- Learns useful features from unlabeled data
- Pretraining tasks include masked token prediction and motif identification.
- Examples: MolBERT, ChemBERTa, GROVER
She pretrained a Transformer to reconstruct masked SMILES tokens — learning hidden structure without needing labeled data.
Reinforcement Learning
- AI agents are rewarded for generating molecules with specific properties.
- Enables multi-objective optimization (potency + safety + synthesizability)
- Models: REINVENT, MolDQN, GCPN
She built an agent that generated new molecules, tweaking structures to maximize binding affinity and minimize toxicity.
Meta & Few-shot Learning
- Tackle low-data scenarios by learning generalizable molecular embeddings.
- Useful for rare diseases or niche therapeutic areas
By teaching models to learn quickly from only a few examples, she opened possibilities for rare disease drug design.
Applications and Success Stories
With weeks of sleepless nights behind her, Ananya’s models were finally maturing. She joined forces with a cross-disciplinary team of medicinal chemists and clinicians, eager to put her pipeline to real-world use. The transition from academic experimentation to impact-driven collaboration was both thrilling and intimidating.
Virtual Screening
AI can rapidly screen millions of virtual compounds to identify potential hits. Platforms like Chemprop and DeepChem have shown strong performance across benchmark datasets.
Ananya’s team used Chemprop to reduce screening time from months to days, identifying leads from millions of candidates.
Antibiotic Discovery
MIT researchers used a VAE (variational autoencoder)-based deep learning system to identify Halicin, a novel antibiotic effective against drug-resistant pathogens (Stokes et al., 2020).
Inspired by MIT, her team used a VAE model to explore uncharted regions of chemical space, leading to a new compound for resistant tuberculosis.
Multi-objective Optimization
AI systems now optimize for multiple objectives simultaneously, including efficacy, ADME properties, and toxicity—an otherwise intractable problem using classical methods.
In Ananya’s case, their pipeline began optimizing molecules for multiple properties at once, balancing efficacy, safety, and manufacturability.
Remaining Challenges
Yet, even in success, the road wasn’t smooth. As the team scaled up their efforts, Ananya discovered cracks in the system. Some predictions defied logic, others couldn’t be explained. She found herself asking: “Can we trust these black boxes?”
| Challenge | Impact |
| Low-quality or biased data | Affects model accuracy and fairness |
| Activity cliffs | Small structural changes with major property shifts |
| Lack of interpretability | Limits trust in predictions |
| Mode collapse in GANs | Reduces the diversity of generated compounds |
Benchmarking platforms like MoleculeNet, MOSES, and GuacaMol help standardize evaluations, keeping her models honest and reproducible.
What Lies Ahead: The Future of Multimodal AI in Drug Discovery
Dr. Ananya didn’t give up. She began looking ahead and learning about exciting new tools. Big AI models, known as foundation models, were now capable of understanding complex biology from vast datasets. These models are trained on large amounts of chemical or biological information and can be fine-tuned for specific tasks like predicting drug effects or generating new compounds. They’re like language models for molecules — powerful, flexible, and reusable.
As her models matured, Ananya tapped into foundation models:
- MolBERT: Pretrained on billions of SMILES strings
- AlphaFold: Protein structure predictor for binding site exploration
- GROVER: Graph-based pretraining for molecule property prediction
These models generalize well across tasks and reduce the need for labeled data.
She also explored federated learning — a technique that allows researchers to train AI models using data from different hospitals or companies without ever moving or sharing the data. Instead, each institution keeps its data locally, and only model updates are shared. This approach protects patient privacy and maintains the confidentiality of sensitive research.
She realized the next chapter wasn’t just about finding new drugs; it was about doing it in a way that’s secure, trustworthy, and fair to everyone.
Therefore, they even began experimenting with federated learning — training on sensitive clinical datasets without transferring patient data.
Conclusion
Years later, Dr. Ananya stood at an international conference stage, presenting a compound that her AI-assisted platform helped bring to Phase I trials. Behind the elegant plots and validation metrics was a story of relentless experimentation, countless lines of code, and a dream to make drug discovery work smarter.
Dr. Ananya’s journey shows how a curious mind, supported by modern AI, can unlock new paths in drug discovery. What started as a failed screen evolved into a multimodal pipeline that fuses biological understanding with computational power.
Multimodal AI is not just a tool — it’s a partner in decoding life’s most complex systems, one data layer at a time.
In this new era, drug discovery is not just faster — it’s smarter, more holistic, and profoundly human-centered.
References
- Deng et al., Artificial Intelligence in Drug Discovery
- Gómez-Bombarelli et al., Design of molecules using VAEs
- Rong et al., Self-Supervised Graph Transformer – GROVER
- Stokes et al., Deep learning for novel antibiotics
Enhance Your AI & ML Skills
Struggling to understand concepts like machine learning, neural networks, or generative AI? Dive deeper into the world of AI with expert-led courses on CloudxLab — your gateway to mastering applied AI for the real world.
Visit CloudxLab

